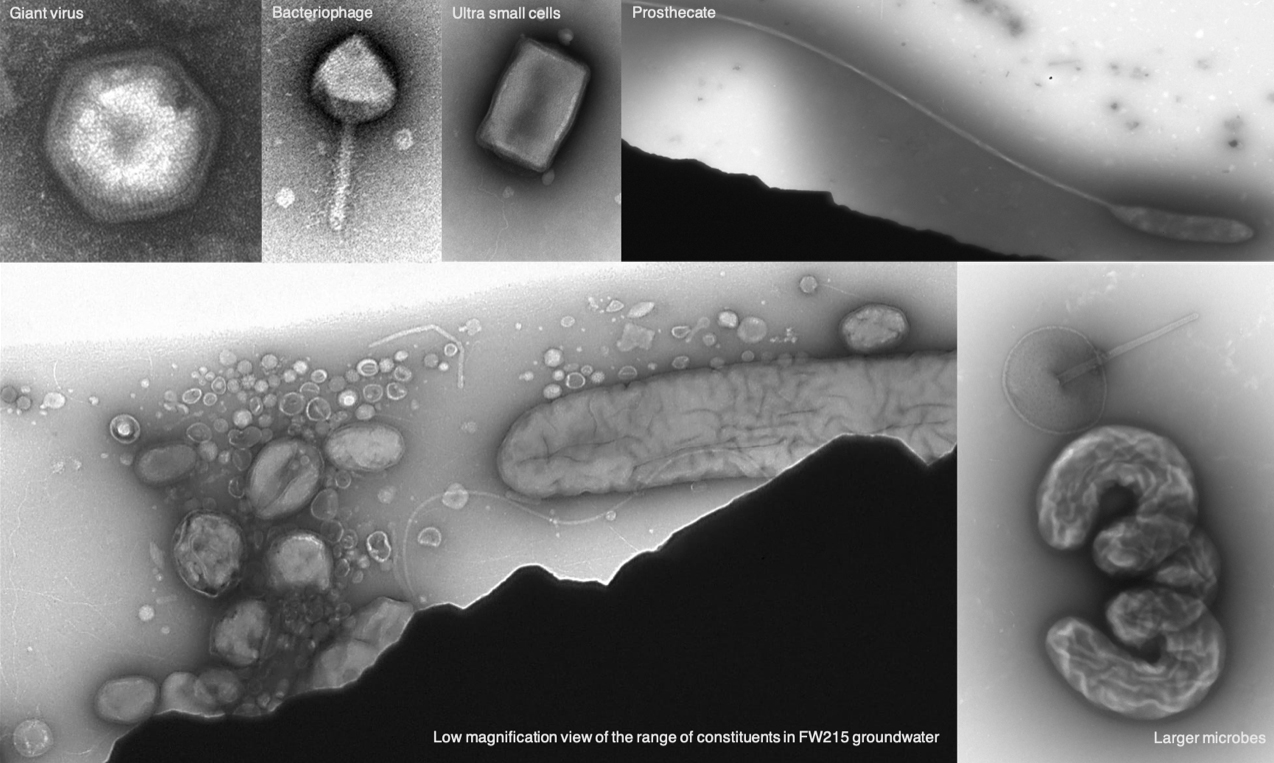
Lab-based ENIGMA researchers enrich, isolate and physiologically/genetically characterize the specific taxa that are predicted to be the most important to the observed field processes. By characterizing genetic functions and linking genotype to phenotype in the laboratory, we can interpret field data mechanistically, leading to pointed hypotheses of the role of particular taxa, proteins, co-factors, and their interactions in creating systems behavior.

Relevant Publications
 Goff Uses KBase to Analyze Genetic Factors Impacting Microbial Fitness in Contaminated Environments - A recent KBase community highlight summarized research accomplishments from Jennifer Goff, who was a postdoctoral researcher with the ENIGMA Science Focus Area in Mike Adams' lab at the University of Georgia (UGA) and is now a professor at SUNY College of Environmental Science and Forestry (ESF). Several of her team’s latest studies, which used KBase to conduct large-scale genetic analyses,… More →
Goff Uses KBase to Analyze Genetic Factors Impacting Microbial Fitness in Contaminated Environments - A recent KBase community highlight summarized research accomplishments from Jennifer Goff, who was a postdoctoral researcher with the ENIGMA Science Focus Area in Mike Adams' lab at the University of Georgia (UGA) and is now a professor at SUNY College of Environmental Science and Forestry (ESF). Several of her team’s latest studies, which used KBase to conduct large-scale genetic analyses,… More → Machine Learning Reveals Patterns of REC Protein Domain Evolution - Bacteria can evolve by exchanging genetic material through a process called recombination. This study used machine learning models combined with functional laboratory experiments to show that after recombination, a bacterial signaling system changed and expanded its protein family, despite considerable selective pressures in place that constrained new modifications. More →
Machine Learning Reveals Patterns of REC Protein Domain Evolution - Bacteria can evolve by exchanging genetic material through a process called recombination. This study used machine learning models combined with functional laboratory experiments to show that after recombination, a bacterial signaling system changed and expanded its protein family, despite considerable selective pressures in place that constrained new modifications. More →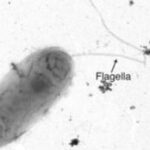 How Bacteria Adapt to Stress - Berkeley Lab researchers uncovered a distinctive adaptation that some bacteria use to quickly form protective communities called biofilms, which help them survive in adverse environments. The work could lead to better stewardship of areas with high levels of heavy metals, nutrients, or other forms of hazardous waste. More →
How Bacteria Adapt to Stress - Berkeley Lab researchers uncovered a distinctive adaptation that some bacteria use to quickly form protective communities called biofilms, which help them survive in adverse environments. The work could lead to better stewardship of areas with high levels of heavy metals, nutrients, or other forms of hazardous waste. More → Huang Explores How Bacteria Store Carbon at 2024 BioEPIC Research SLAM - Congratulations to Joshua Jiaqi Huang, a UC Berkeley graduate student researcher in Biosciences senior faculty scientist Adam Arkin’s group, who represented ENIGMA and Biosciences at the 2024 BioEPIC Research SLAM. More →
Huang Explores How Bacteria Store Carbon at 2024 BioEPIC Research SLAM - Congratulations to Joshua Jiaqi Huang, a UC Berkeley graduate student researcher in Biosciences senior faculty scientist Adam Arkin’s group, who represented ENIGMA and Biosciences at the 2024 BioEPIC Research SLAM. More → fast.genomics: A Fast Comparative Genome Browser for Diverse Bacteria and Archaea - ENIGMA researchers have developed fast.genomics, a new tool which allows users to search for a protein and identify other proteins that share a common ancestor or are located in similar genomic regions. More →
fast.genomics: A Fast Comparative Genome Browser for Diverse Bacteria and Archaea - ENIGMA researchers have developed fast.genomics, a new tool which allows users to search for a protein and identify other proteins that share a common ancestor or are located in similar genomic regions. More → Multi-disciplinary research develops a deeper understanding of subsurface microbiology - ENIGMA is organized into three aims that reflect the different research scales (i.e., field level to molecular level) required to build a comprehensive picture of subsurface microbiology at the Oak Ridge Reservation (ORR): Subsurface Observatory, Environmental Atlas, and Environmental Simulations. Several key research initiatives, described in this cross-aim overview, highlight this framework in action. More →
Multi-disciplinary research develops a deeper understanding of subsurface microbiology - ENIGMA is organized into three aims that reflect the different research scales (i.e., field level to molecular level) required to build a comprehensive picture of subsurface microbiology at the Oak Ridge Reservation (ORR): Subsurface Observatory, Environmental Atlas, and Environmental Simulations. Several key research initiatives, described in this cross-aim overview, highlight this framework in action. More →