The Science
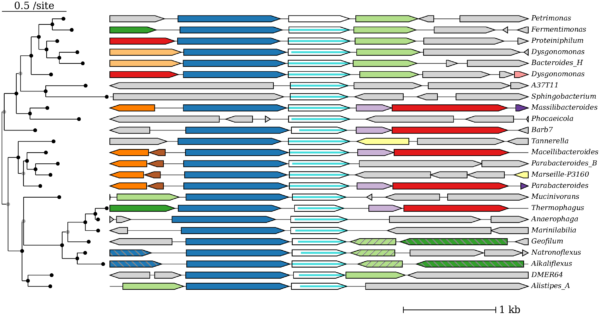
While scientists have sequenced genomes for over ten thousand species of bacteria and archaea, tools for browsing these genomes are cumbersome. A key challenge is that there is no convenient way to explore which sequenced proteins are related to a given protein of interest. ENIGMA researchers developed a web-based tool, fast.genomics, which uses accelerated searches to find similar proteins and to view the results. In particular, fast.genomics shows which groups of bacteria or archaea contain similar proteins and which other proteins, if any, are conserved across multiple species along with the protein of interest.
The Impact
Fast.genomics makes it much quicker and easier to find conserved gene neighbors. Because conserved neighbors often have related functions, this can help predict the function of the protein. Similarly, if two proteins have related functions, then they often appear together across genomes. Fast.genomics has a quick tool to compare the presence and absence of two proteins. Overall, fast.genomics makes it easier to use sequenced genomes to predict protein function.
Summary
Genome sequencing has revealed an incredible diversity of bacteria and archaea, but there are no fast and convenient tools for browsing across these genomes. It is cumbersome to view the prevalence of homologs, or genes descended from a common ancestor, for a protein of interest, or assess whether a protein or its homologs are co-located with other similar proteins across many prokaryotic species. ENIGMA web-based tool, fast.genomics, uses two strategies to support fast browsing across the diversity of prokaryotes. First, the database of genomes is split up. The main database contains one representative from each of the 6,377 genera that have a high-quality genome, and additional databases for each taxonomic order contain up to ten representatives of each species. Second, homologs of proteins of interest are identified quickly by using accelerated searches, usually in a few seconds. Once homologs are identified, fast.genomics can quickly show their incidence across taxa, view their neighboring genes, or compare the frequency of two different proteins. Fast.genomics is available at https://fast.genomics.lbl.gov.
Contact
Morgan Price, Computational Biologist Research Scientist
Lawrence Berkeley National Laboratory
mnprice@lbl.gov
Funding
This material by ENIGMA (Ecosystems and Networks Integrated with Genes and Molecular Assemblies), a Science Focus Area Program at Lawrence Berkeley National Laboratory, is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research under contract number DE-AC02-05CH11231.
Publication
Price, M.N.; Arkin, A.P. (2024) A fast comparative genome browser for diverse bacteria and archaea. PLoS One, [DOI]: https://doi.org/10.1371/journal.pone.0301871. OSTI:2335832
