 Microbial Remains Shape Community Composition and Growth of Soil Microorganisms - The Science Microbes, which include organisms like bacteria, are tiny living things that inhabit virtually every environment on Earth. Soil microbes play a crucial role in breaking down organic matter and are crucial drivers of global nutrient cycles that support all terrestrial ecosystems. ENIGMA scientists studied what happens… News Article →
Microbial Remains Shape Community Composition and Growth of Soil Microorganisms - The Science Microbes, which include organisms like bacteria, are tiny living things that inhabit virtually every environment on Earth. Soil microbes play a crucial role in breaking down organic matter and are crucial drivers of global nutrient cycles that support all terrestrial ecosystems. ENIGMA scientists studied what happens… News Article →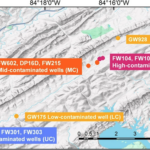 Functional Resilience in Microbes found in Groundwater - ENIGMA researchers studied microbes living in groundwater at a site contaminated with metal, acid, and other waste. They compared wells with high, medium, low, and no levels of contamination. They found that even when microbial species count decreased under stress, groups of microbes compensated to sustain essential functions. News Article →
Functional Resilience in Microbes found in Groundwater - ENIGMA researchers studied microbes living in groundwater at a site contaminated with metal, acid, and other waste. They compared wells with high, medium, low, and no levels of contamination. They found that even when microbial species count decreased under stress, groups of microbes compensated to sustain essential functions. News Article → Goff Uses KBase to Analyze Genetic Factors Impacting Microbial Fitness in Contaminated Environments - A recent KBase community highlight summarized research accomplishments from Jennifer Goff, who was a postdoctoral researcher with the ENIGMA Science Focus Area in Mike Adams' lab at the University of Georgia (UGA) and is now a professor at SUNY College of Environmental Science and Forestry (ESF). Several of… News Article →
Goff Uses KBase to Analyze Genetic Factors Impacting Microbial Fitness in Contaminated Environments - A recent KBase community highlight summarized research accomplishments from Jennifer Goff, who was a postdoctoral researcher with the ENIGMA Science Focus Area in Mike Adams' lab at the University of Georgia (UGA) and is now a professor at SUNY College of Environmental Science and Forestry (ESF). Several of… News Article →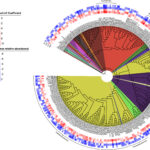 Adding Nutrients to Groundwater Yields Reproducible Response in Nearby Microbial Communities - Emulsified vegetable oil injections into groundwater promoted reproducible geochemical and microbial changes across an eight-year time gap. News Article →
Adding Nutrients to Groundwater Yields Reproducible Response in Nearby Microbial Communities - Emulsified vegetable oil injections into groundwater promoted reproducible geochemical and microbial changes across an eight-year time gap. News Article → Machine Learning Reveals Patterns of REC Protein Domain Evolution - Bacteria can evolve by exchanging genetic material through a process called recombination. This study used machine learning models combined with functional laboratory experiments to show that after recombination, a bacterial signaling system changed and expanded its protein family, despite considerable selective pressures in place that constrained new modifications. News Article →
Machine Learning Reveals Patterns of REC Protein Domain Evolution - Bacteria can evolve by exchanging genetic material through a process called recombination. This study used machine learning models combined with functional laboratory experiments to show that after recombination, a bacterial signaling system changed and expanded its protein family, despite considerable selective pressures in place that constrained new modifications. News Article →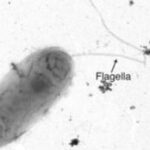 How Bacteria Adapt to Stress - Berkeley Lab researchers uncovered a distinctive adaptation that some bacteria use to quickly form protective communities called biofilms, which help them survive in adverse environments. The work could lead to better stewardship of areas with high levels of heavy metals, nutrients, or other forms of hazardous waste. News Article →
How Bacteria Adapt to Stress - Berkeley Lab researchers uncovered a distinctive adaptation that some bacteria use to quickly form protective communities called biofilms, which help them survive in adverse environments. The work could lead to better stewardship of areas with high levels of heavy metals, nutrients, or other forms of hazardous waste. News Article → From Field-to-Lab and Lab-to-Field: How Microbes Evolve, Form Partnerships and Work Together - Researchers in the Institute for Systems Biology’s Baliga Lab examined model organisms from two classes of microbes whose interaction converts over a gigaton of carbon to methane each year and found that gene mutations selected for over a relatively short timeframe in the two microbes –Desulfovibrio vulgaris (Dv)… News Article →
From Field-to-Lab and Lab-to-Field: How Microbes Evolve, Form Partnerships and Work Together - Researchers in the Institute for Systems Biology’s Baliga Lab examined model organisms from two classes of microbes whose interaction converts over a gigaton of carbon to methane each year and found that gene mutations selected for over a relatively short timeframe in the two microbes –Desulfovibrio vulgaris (Dv)… News Article → Huang Explores How Bacteria Store Carbon at 2024 BioEPIC Research SLAM - Congratulations to Joshua Jiaqi Huang, a UC Berkeley graduate student researcher in Biosciences senior faculty scientist Adam Arkin’s group, who represented ENIGMA and Biosciences at the 2024 BioEPIC Research SLAM. News Article →
Huang Explores How Bacteria Store Carbon at 2024 BioEPIC Research SLAM - Congratulations to Joshua Jiaqi Huang, a UC Berkeley graduate student researcher in Biosciences senior faculty scientist Adam Arkin’s group, who represented ENIGMA and Biosciences at the 2024 BioEPIC Research SLAM. News Article → fast.genomics: A Fast Comparative Genome Browser for Diverse Bacteria and Archaea - ENIGMA researchers have developed fast.genomics, a new tool which allows users to search for a protein and identify other proteins that share a common ancestor or are located in similar genomic regions. News Article →
fast.genomics: A Fast Comparative Genome Browser for Diverse Bacteria and Archaea - ENIGMA researchers have developed fast.genomics, a new tool which allows users to search for a protein and identify other proteins that share a common ancestor or are located in similar genomic regions. News Article → Jinwoo Im is ENIGMA’s Third BioEPIC Virtual Research Slam Presenter - With anticipated occupancy early 2025, Berkeley Lab’s newest building Biological and Environmental Program Integration Center (BioEPIC), is located in the beautiful Berkeley hills overlooking San Francisco Bay and the Golden Gate Bridge. As part of Berkeley Lab’s Earth Month celebrations, a Research SLAM was hosted by Biosciences (BSA)… News Article →
Jinwoo Im is ENIGMA’s Third BioEPIC Virtual Research Slam Presenter - With anticipated occupancy early 2025, Berkeley Lab’s newest building Biological and Environmental Program Integration Center (BioEPIC), is located in the beautiful Berkeley hills overlooking San Francisco Bay and the Golden Gate Bridge. As part of Berkeley Lab’s Earth Month celebrations, a Research SLAM was hosted by Biosciences (BSA)… News Article → Multi-disciplinary research develops a deeper understanding of subsurface microbiology - ENIGMA is organized into three aims that reflect the different research scales (i.e., field level to molecular level) required to build a comprehensive picture of subsurface microbiology at the Oak Ridge Reservation (ORR): Subsurface Observatory, Environmental Atlas, and Environmental Simulations. Several key research initiatives, described in this cross-aim… News Article →
Multi-disciplinary research develops a deeper understanding of subsurface microbiology - ENIGMA is organized into three aims that reflect the different research scales (i.e., field level to molecular level) required to build a comprehensive picture of subsurface microbiology at the Oak Ridge Reservation (ORR): Subsurface Observatory, Environmental Atlas, and Environmental Simulations. Several key research initiatives, described in this cross-aim… News Article →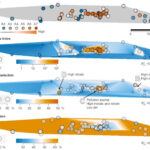 Nuclear Waste Sites Yield Microbial Ecosystem Insights - In a flagship seven-year study, published this January in the journal Nature Microbiology, ENIGMA researchers explored how environmental stresses influence the composition and structure of microbial communities in the groundwater of the Oak Ridge Reservation (ORR), a former nuclear waste disposal site. News Article →
Nuclear Waste Sites Yield Microbial Ecosystem Insights - In a flagship seven-year study, published this January in the journal Nature Microbiology, ENIGMA researchers explored how environmental stresses influence the composition and structure of microbial communities in the groundwater of the Oak Ridge Reservation (ORR), a former nuclear waste disposal site. News Article →