 University of Oklahoma
University of Oklahoma
Principal Investigator
jzhou@ou.edu
(405) 325-6073
https://www.ou.edu/ieg
Jizhong Zhou has expertise in molecular biology, microbial genomics, microbial ecology, molecular evolution, theoretical ecology, metagenomics, and genomic technologies, as well as array-based bioinformatics for environmental studies. He has pioneered the development of array-based genomic technologies for environmental studies. His lab does much of the field sample processing for sequencing and as such is involved in multiple collaborations across the three aims where their packages/pipelines, mechanisms, and predictions of the microbiome and biogeochemical dynamics are used.
He provides high-quality microbiome data to support the field and laboratory research, including amplicon sequencing of phylogenetic marker genes (e.g. 16S, 18S) and key functional genes (e.g. amoA), functional gene microarray test. He develops and applies statistical tools (particularly for network analysis, time series analysis, community assembly mechanisms), ecological models, and omics-enabled biogeochemical models to approach a mechanistic and predictive understanding of the subsurface microbiome dynamics and their functioning in biogeochemical processes.
Relevant Publications
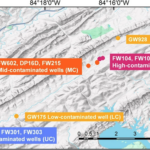 Functional Resilience in Microbes found in Groundwater - ENIGMA researchers studied microbes living in groundwater at a site contaminated with metal, acid, and other waste. They compared wells with high, medium, low, and no levels of contamination. They found that even when microbial species count decreased under stress, groups of microbes compensated to sustain essential functions. More →
Functional Resilience in Microbes found in Groundwater - ENIGMA researchers studied microbes living in groundwater at a site contaminated with metal, acid, and other waste. They compared wells with high, medium, low, and no levels of contamination. They found that even when microbial species count decreased under stress, groups of microbes compensated to sustain essential functions. More →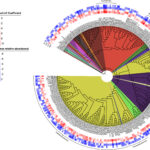 Adding Nutrients to Groundwater Yields Reproducible Response in Nearby Microbial Communities - Emulsified vegetable oil injections into groundwater promoted reproducible geochemical and microbial changes across an eight-year time gap. More →
Adding Nutrients to Groundwater Yields Reproducible Response in Nearby Microbial Communities - Emulsified vegetable oil injections into groundwater promoted reproducible geochemical and microbial changes across an eight-year time gap. More → Multi-disciplinary research develops a deeper understanding of subsurface microbiology - ENIGMA is organized into three aims that reflect the different research scales (i.e., field level to molecular level) required to build a comprehensive picture of subsurface microbiology at the Oak Ridge Reservation (ORR): Subsurface Observatory, Environmental Atlas, and Environmental Simulations. Several key research initiatives, described in this cross-aim overview, highlight this framework in action. More →
Multi-disciplinary research develops a deeper understanding of subsurface microbiology - ENIGMA is organized into three aims that reflect the different research scales (i.e., field level to molecular level) required to build a comprehensive picture of subsurface microbiology at the Oak Ridge Reservation (ORR): Subsurface Observatory, Environmental Atlas, and Environmental Simulations. Several key research initiatives, described in this cross-aim overview, highlight this framework in action. More →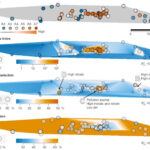 Nuclear Waste Sites Yield Microbial Ecosystem Insights - In a flagship seven-year study, published this January in the journal Nature Microbiology, ENIGMA researchers explored how environmental stresses influence the composition and structure of microbial communities in the groundwater of the Oak Ridge Reservation (ORR), a former nuclear waste disposal site. More →
Nuclear Waste Sites Yield Microbial Ecosystem Insights - In a flagship seven-year study, published this January in the journal Nature Microbiology, ENIGMA researchers explored how environmental stresses influence the composition and structure of microbial communities in the groundwater of the Oak Ridge Reservation (ORR), a former nuclear waste disposal site. More →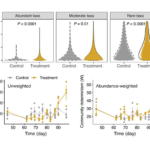 New Platform Explores Ecological Drivers of Change in Microbial Communities - ENIGMA researchers assess mechanisms for microbial taxa and community dynamics using process models. More →
New Platform Explores Ecological Drivers of Change in Microbial Communities - ENIGMA researchers assess mechanisms for microbial taxa and community dynamics using process models. More →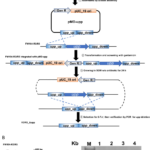 Genome Editing in Rhodanobacter denitrificans - ENIGMA researchers demonstrated the development and application of a markerless deletion mutagenesis system in nitrate-reducing bacterium Rhodanobacter denitrificans. This method marks a crucial step in advancing Rhodanobacter as a model denitrifying bacterium for the study of denitrification in groundwater ecosystems and diverse molecular mechanisms of low-pH resistance. More →
Genome Editing in Rhodanobacter denitrificans - ENIGMA researchers demonstrated the development and application of a markerless deletion mutagenesis system in nitrate-reducing bacterium Rhodanobacter denitrificans. This method marks a crucial step in advancing Rhodanobacter as a model denitrifying bacterium for the study of denitrification in groundwater ecosystems and diverse molecular mechanisms of low-pH resistance. More →