Adaptation to previous stressors influence which genes acquire mutations during experimental evolution
M.L. Kempher, X. Tao, R. Song, B. Wu, D.A. Stahl, J.D. Wall, A.P. Arkin, A. Zhou, J. Zhou. “Effects of genetic and physiological divergence on the evolution of a sulfate-reducing bacterium under conditions of elevated temperature.” mBio, 11(4), e00569-20 (2020). [doi: 10.1128/mBio.00569-20]
The Science
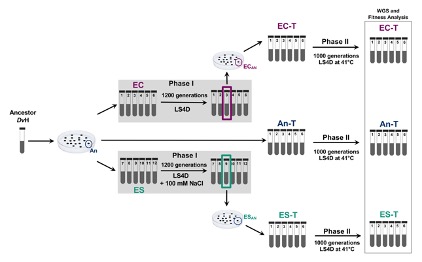
It has long been known that the process of natural selection will favor traits that are beneficial to the survival of the organism. However, how other influences, such as the history of the organism, influences adaptation to new environmental conditions has been difficult to determine. We established a two phase experimental design to evaluate how three groups of populations of a sulfate reducing bacterium with different histories adapted to elevated temperature. We determined that all the populations adapted to similar fitness levels, but history affected the genetic changes acquired by each population.
The Impact
Understanding what influences an evolutionary trajectory has been difficult for researchers to study given the time scale required to typically follow multiple generations of higher organisms. However, microorganisms grow and adapt quickly which provides scientists an opportunity to study evolution in real time in the laboratory. Using microbial evolution systems, we are able to evaluate if different life histories affect how an organism adapts to a new and/or stressful environment.
Summary
Determining if and how differences in history influence adaptation has been a longstanding goal of evolutionary biologists. To evaluate this in the laboratory, we experimentally evolved three groups of six populations of the sulfate reducing bacterium, Desulfovibrio vulgaris, for 1,000 generations under elevated temperature. Two of the groups had diverged during a previous study while adapting to elevated salt conditions or control, nonstress conditions for 1,200 generations. The third group had not experienced any experimental evolution and was included as a control. All eighteen populations adapted to the elevated temperature which was evidenced by faster growth rates and improved fitness compared to the ancestor. Whole genome resequencing revealed that the underlying mutations were quite different between the two groups with different evolutionary histories. The populations from each group adopted different functional strategies to adapt to increased temperature indicating that the historical environment significantly influenced evolution. Improving our understanding of the driving forces of evolution will help us to predict how microbes will respond and adapt to various environmental stressors.
Contact
Megan Kempher
University of Oklahoma
kempher@ou.edu
Jizhong Zhou
University of Oklahoma
jzhou@ou.edu
