Modeling microbial interactions in synthetic communities offers insights into environmental processes
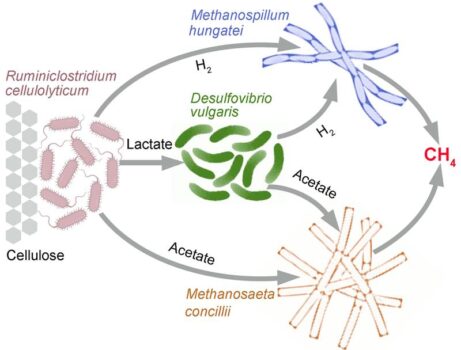
Synthetic microbial communities are a powerful tool to study microbial interactions. However, most studies to date have focused on binary interactions between only two microbial species to describe their interactions and were constrained by individual methods. This study investigated complex interactions among four cross-feeding microorganisms in a synthetic community (SynCom) that converts cellulose to methane and carbon dioxide. The researchers used the SynCom as a model system for studying the biochemical and physiological responses and function changes of microbial community behavior in estuarine wetlands under the threat of seawater intrusion. Scientists expect sea level rise will bring greater levels of seawater intrusion into coastal wetlands and aquifers. The inclusion of a sulfate-reducing bacterium in the synthetic community enabled the researchers to examine the impact of sulfate intrusion on community dynamics.
The researchers characterized the biochemical and physiological responses of each microorganism in the synthetic community via proteomic analysis to study their ecological and metabolic responses and to model the inter-species interactions governing carbon exchange. Stoichiometric modeling indicated that sulfate addition changed carbon and electron fluxes, as well as metabolic interactions, in predicted ways, but also revealed positive and negative synergies as emergent properties of high-order microbial interactions. This study paves the way toward a more predictive understanding of the impact of environmental perturbations on microbial interactions sustaining geochemically significant processes in natural systems.
Contact
Chongle Pan
University of Oklahoma
cpan@ou.edu
Funding
This study is primarily funded by the Genome Sciences Program of the Office of Biological and Environmental Research under project FWP SCW1677. Part of the metabolic modeling was supported by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA; http://enigma.lbl.gov), a Science Focus Area Program at Lawrence Berkeley National Laboratory that is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research, under contract number DE-AC02-05CH11231. Part of the proteomics analysis was supported by the IDeA National Resource for Quantitative Proteomics and a National Institutes of Health grant (R01AT011618). The contributions of M.J.M. and N.Q.W. were supported by a National Science Foundation grant (1515843). The publication fee was supported by the Open Access Fund of the University of Oklahoma Libraries.
Publications
Wang. D, K. A. Hunt, P. Candry, X. Tao, N. Q. Wofford, J. Zhou, M. J. McInerney, D. A. Stahl, R. S. Tanner, A. Zhou, M. Winkler and C. Pan (2023). “Cross-Feedings, Competition, and Positive and Negative Synergies in a Four-Species Synthetic Community for Anaerobic Degradation of Cellulose to Methane.” Mbio: e03189-03122. [DOI: https://doi.org/10.1128/mbio.03189-22]
