Tools to map target genes of bacterial two-component system response regulators
Rajeev L, Garber ME, Mukhopadhyay, A
Environmental Microbiology Reports. 2020 doi.org/10.1111/1758-2229.12838
Microbial signaling and gene regulation is an important aspect of the ecology being studied by the ENIGMA project at the Oakridge Field Site. 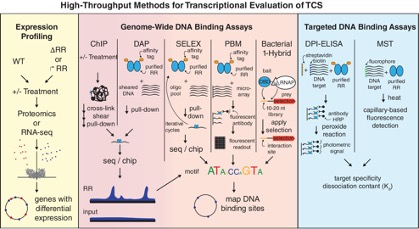 There are several methods currently available to examine transcriptionally acting signaling systems in bacteria. Approaches such as DAP-seq, an emerging method, have their origins in the DAP-chip approach, a legacy of earlier ENIGMA studies. Here we showcase DAP-chip, which was used to examine all two-component signaling systems in the model sulfate reducer, Desulfovibrio vulgaris. Recently, DAP-seq was used to examine Cu and Zn regulating systems in the P. stutzeri
There are several methods currently available to examine transcriptionally acting signaling systems in bacteria. Approaches such as DAP-seq, an emerging method, have their origins in the DAP-chip approach, a legacy of earlier ENIGMA studies. Here we showcase DAP-chip, which was used to examine all two-component signaling systems in the model sulfate reducer, Desulfovibrio vulgaris. Recently, DAP-seq was used to examine Cu and Zn regulating systems in the P. stutzeri
