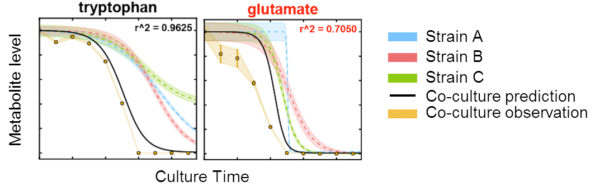
 Exometabolomic profiling was used to examine the time-varying substrate depletion from a mixture of 19 amino acids and glucose by two Pseudomonads and one Bacillus FRC isolates. ENIGMA researchers at Lawrence Berkeley National Lab examined if the first substrates depleted resulted in maximal growth rate, or related to growth medium or biomass composition and found surprisingly few correlations. We also modeled patterns of substrate depletion, and these models were used to examine if substrate usage preferences and substrate depletion kinetics of three microbial isolates can be used to predict the metabolism of the pooled isolates in co-culture. We found that most of the substrates fit the model predictions, indicating that the microbes are not altering their behaviors for these substrates in the presence of competitors. Compounds that deviate from the model highlight substrates that could be involved in species-species interactions within the consortium.
Exometabolomic profiling was used to examine the time-varying substrate depletion from a mixture of 19 amino acids and glucose by two Pseudomonads and one Bacillus FRC isolates. ENIGMA researchers at Lawrence Berkeley National Lab examined if the first substrates depleted resulted in maximal growth rate, or related to growth medium or biomass composition and found surprisingly few correlations. We also modeled patterns of substrate depletion, and these models were used to examine if substrate usage preferences and substrate depletion kinetics of three microbial isolates can be used to predict the metabolism of the pooled isolates in co-culture. We found that most of the substrates fit the model predictions, indicating that the microbes are not altering their behaviors for these substrates in the presence of competitors. Compounds that deviate from the model highlight substrates that could be involved in species-species interactions within the consortium.
Significance
- Time-resolved resource usage of strains grown alone was used to formulate models for substrate usage in a co-culture setting, assuming no overt competition between the strains for the substrate
- The model accurately predicted mixed-culture substrate depletion for about half of the resources provided in the medium
- Deviation from the models indicate resources of interest to study further
- This study serves as a proof-of-principle for a method to identify critical resources in a more complex environment
