Alkaline-SDS cell lysis of microbes with acetone precipitation for proteomic sample preparation in 96-well plate format
The Science
The nutrient availability and complexity of environmental perturbations are important drivers of microbiome dynamics and activities. In terrestrial subsurface environments such as at the ENIGMA ORR, new microbial isolates and microbial communities are discovered rapidly. To understand the physiological, structural, and dynamic changes of these isolates in defined lab conditions, hundreds of samples are generated for proteomic analysis. By conducting laboratory tests on plate-based proteomic sample preparation protocols, ENIGMA researchers established an automated protocol that enables fast, low cost, and low-variance proteomics sample preparation to analyze synthetic microbial communities.
The Impact
This protocol, combined with previously established automated protein quantification and protein normalization protocols, provides a rapid, cost-effective method to prepare LC-MS proteomic samples from bacteria and non-filamentous fungi cell cultures. The flexible design of this protocol enables one researcher to prepare thousands of bottom-up proteomic samples per week. This work supports the integration of complementary technologies to achieve many core ENIGMA scientific aims.
Summary
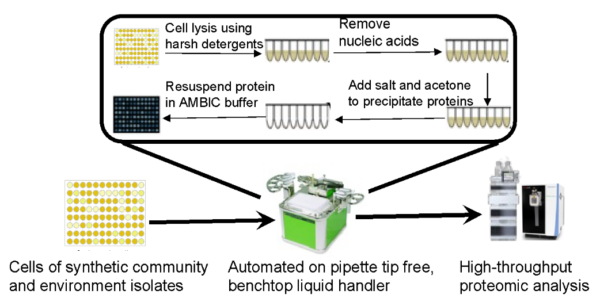
ENIGMA researchers detailed a step-by-step protocol that consists of cell lysis in alkaline chemical buffer (NaOH/SDS) followed by protein precipitation with high-ionic strength acetone in 96-well format. The protocol works for a broad range of microbes (e.g., Gram-negative bacteria, Gram-positive bacteria, non-filamentous fungi) and the resulting proteins are ready for tryptic digestion for bottom-up quantitative proteomic analysis without the need for additional cleanup procedures. The yield of protein using this protocol increases linearly with respect to the amount of starting biomass. Using a benchtop automated liquid dispenser, a cost-effective and environmentally-friendly option for eliminating pipette tips and reducing reagent waste, the protocol takes approximately 30 minutes to extract protein from 96 samples. Tests on mock mixtures showed that the biomass composition structure closely resembles the experimental design. Lastly, researchers applied the protocol to composition analysis of a synthetic community of environmental isolates grown on two different media. This protocol has been developed to facilitate rapid, low-variance sample preparation of hundreds of samples and allow flexibility for future protocol development.

Lawrence Berkeley National Laboratory
Contact
Christopher Petzold, Staff Scientist
Biological Systems & Engineering Division
Lawrence Berkeley National Laboratory
Berkeley CA 94720
cjpetzold@lbl.gov

Lawrence Berkeley National Laboratory
Yan Chen, Project Scientist
Biological Systems & Engineering Division
Lawrence Berkeley National Laboratory
Berkeley CA 94720
yanchen1998@lbl.gov
Funding
This material by ENIGMA- Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Science Focus Area Program at Lawrence Berkeley National Laboratory is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research under contract number DE-AC02-05CH11231.
Publications
Chen, Y.; J.W. Gin, Y. Wang, M. de Raad, S. Tan, N.J. Hillson, T.R. Northen, P.D. Adams and C.J. Petzold (2023) Alkaline-SDS cell lysis of microbes with acetone protein precipitation for proteomic sample preparation in 96-well plate format. Plos One. [doi]:10.1371/journal.pone.0288102 {PMID}:37418444 PMCID:PMC10328223 OSTI: 1988658
