ENIGMA researchers assess mechanisms for microbial taxa and community dynamics using process models.
The Science
A microbial community usually has hundreds or thousands of species. The individual number of each species generally keeps changing over time due to various ecological processes, such as environment selection, resource competition, immigration, and random birth or death. Some processes are species-specific, while others are more random. ENIGMA scientists developed different models to predict the changes caused by different types of ecological processes. Integrating two distinct model types revealed the drivers of change across assorted species within the same community, and resulted in better predictions.
The Impact
One fundamental goal in microbial ecology is to predict how microbial diversity changes across space and time. Although spatial patterns of microbial communities have recently received attention, microbial temporal dynamics remain poorly understood, primarily due to the lack of appropriate experimental data and theoretical frameworks. By reconciling niche and neutral perspectives, this study developed a novel process model‐based framework to effectively encapsulate microbial species temporal dynamics which is powerful for quantitatively assessing the assembly mechanisms underlying microbial community dynamics. This study represents a significant advance in explaining microbial temporal dynamics toward predictive microbial community ecology.
Summary
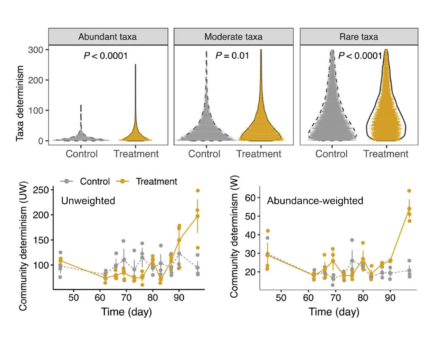
Disentangling the assembly mechanisms controlling community composition, structure, distribution, functions, and dynamics is a central issue in ecology. Although various approaches have been proposed to examine community assembly mechanisms, quantitative characterization is challenging, particularly in microbial ecology. Here, ENIGMA researchers present a novel approach for quantitatively delineating community assembly mechanisms by combining the consumer–resource model with a neutral model in stochastic differential equations. Using time-series data from anaerobic bioreactors that target microbial 16S rRNA genes, researchers tested the applicability of three ecological models: the consumer–resource model, the neutral model, and the combined model. Their results revealed that model performances varied substantially as a function of population abundance and/or process conditions. The combined model performed best for abundant taxa in the treatment bioreactors where process conditions were manipulated. In contrast, the neutral model showed the best performance for rare taxa. Further analysis indicated that immigration rates decreased with taxa abundance and competitions between taxa were strongly correlated with phylogeny, but only within a certain phylogenetic distance. The determinism underlying taxa and community dynamics were quantitatively assessed, showing greater determinism in the treatment bioreactors that aligned with the subsequent abnormal system functioning.
Contact

Jizhong Zhou, George Lynn Cross Research Professor; Director, Institute for Environmental Genomics (IEG)
Department of Microbiology & Plant Biology, University of Oklahoma
jzhou@ou.edu
Funding
The initial development of the theoretical framework for modeling species dynamics Phase I was supported by the National Science Foundation of China to Y. Y. The further improvement of the theoretical framework for assessing community-level determinism Phase II was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research DOE-BER; also in part through ENIGMA- Ecosystems and Networks Integrated with Genes and Molecular Assemblies, a Science Focus Area at Lawrence Berkeley National Laboratory, supported by DOE-BER; and by the U.S. National Science Foundation. The maintenance of the bioreactors was partly supported by the U.S. Environmental Protection Agency and the Science Alliance—Tennessee Center of Excellence. The China Scholarship Council CSC provided support for L. W., Q. G., and H. Y.
Publication
Wu L.; Y. Yang, D. Ning, Q. Gao, H. Yin, N. Xiao, B.Y. Zhou, S. Chen, Q. He, J. Zhou (2023) Assessing Mechanisms for Microbial Taxa and Community Dynamics Using Process Models. mLife. [DOI]:10.1002/mlf2.12076. OSTI: 2205081
