ENIGMA researchers at University of Oklahoma, Lawrence Berkeley Lab, University of Washington, and University of Missouri reveal that the accumulation and induction of key cellular components persist in bacteria for greater than 5000 generations. However, the physiological and transcriptional responses to high salinity are altered. The mechanistic changes in evolved genotypes suggested that growth energy efficiency might be a key factor for selection. Aifen Zhou is interested in the interaction between bacteria and the environment; here, the co-existence of sulfate-reducing bacterium (SRB) and high salinity in natural habitats and heavy metal contaminated field sites laid the foundation for the study of the salt adaptation of DvH, a model SRB, with experimental evolution.
Questions addressed
- What is the major driving force for salt adaptation in DvH over a long-term experimental evolution?
- How did the cellular components such as metabolites and phospholipid fatty acids (PLFAs) change over long term evolution?
Major findings
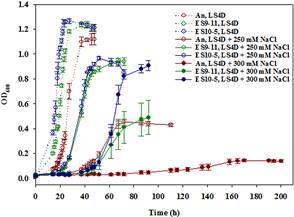
Comparison between the best-adapted clones ES10-5 and ES9-11, which had evolved under salt stress for 5,000 and 1,200 generations, respectively, demonstrated the continuous improvement of salt tolerance; glutamate as a key osmolyte and i17:1ω9c as the major PLFA for salt tolerance in DvH; growth energy efficiency might be a key factor for selection; there is a restorative trend of metabolic, physiological, and transcriptional changes in the long-term evolution of DvH; the relationship between a genotype and a phenotype is very complex.
Rapid genetic and phenotypic adaptation of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough to salt stress was observed during experimental evolution. In order to identify key metabolites important for salt tolerance, a clone, ES10-5, isolated from population ES10 and allowed to experimentally evolve under salt stress for 5,000 generations, was analyzed and compared to clone ES9-11, which was isolated from population ES9 and had evolved under the same conditions for 1,200 generations. These two clones were chosen because they represented the best-adapted clones among six independently evolved populations. ES10-5 acquired new mutations in genes potentially involved in salt tolerance, in addition to the preexisting mutations and different mutations in the same genes as in ES9-11. Most basal abundance changes of metabolites and phospholipid fatty acids (PLFAs) were lower in ES10-5 than ES9-11, but an increase of glutamate and branched PLFA i17:1ω9c under high salinity was persistent. ES9-11 had decreased cell motility compared to the ancestor; in contrast, ES10-5 showed higher cell motility under both nonstress and high salinity conditions. Both genotypes displayed better growth energy efficiencies than the ancestor under nonstress or high salinity. Consistently, ES10-5 did not display most of the basal transcriptional changes observed in ES9-11. Still, it showed increased expression of genes involved in glutamate biosynthesis, cation efflux, and energy metabolism under high salinity. These results demonstrated the role of glutamate as a key osmolyte and i17:1ω9c as the major PLFA for salt tolerance in D. vulgaris.
DOI: 10.1128/mBio.01780-1714 November 2017 mBio vol. 8no. 6 e01780-17
Key Metabolites and Mechanistic Changes for Salt Tolerance in an Experimentally Evolved Sulfate-Reducing Bacterium, Desulfovibrio vulgaris.
Aifen Zhoua, Rebecca Laub, Richard Baranb, Jincai Maa, Frederick von Netzerc, Weiling Shia, Drew Gorman-Lewisd, Megan L. Kemphera, Zhili Hea, Yujia Qina, Zhou Shia, Grant M. Zanee, Liyou Wua, Benjamin P. Bowenb, Trent R. Northenb, Kristina L. Hilleslandf, David A. Stahlc, Judy D. Walle, Adam P. Arkinb,g, Jizhong Zhoua,h,i
