
Kothari, Ankita.; S. Roux, H. Zhang, A. Prieto, J-M Chandonia, S. Spencer, X. Wu, A M. Deutschbauer, A. P. Arkin, E J. Alm, R. Chakraborty, A Mukhopadhyay (2021) Ecogenomics of groundwater viruses suggests niche differentiation linked to specific environmental tolerance. mSystems [doi]:1128/mSystems.00537-21 {PMID}: 34184913 PMCID: PMC8269241 OSTI:1807913
Niche Specific Genes from Phage Genomes are Advantageous for Communities of Groundwater Bacteria
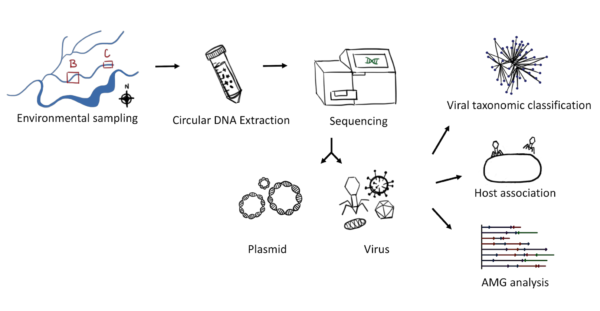
Ground water ecosystems contain microbes as well as mobile genetic elements that originate from microbes. We examined these extra chromosomal DNA in several ground water systems from background and contaminated sites at the Oak Ridge Reservation (ORR) in Tennessee, focusing on selectively isolating and sequencing them. In our study, we analyzed plasmidome sequence information for the presence of phage/viral genomes. Using data from seven ground water samples, we identified 200 viral sequences, which clustered into 41 ~ genus-level viral clusters (equivalent to viral genera) including 9 known and 32 possible new genera. Then, to assess potential bacterial hosts for these phages, we used publicly available bacterial genome sequences and 261 bacterial isolates from this ORR field site. We were able to predict bacterial hosts for 76 of the 200 viral sequences to a range of bacterial phyla. Most notably, we found that these viral genomes encoded functions such as metal tolerance, which is consistent with the metal contamination observed in the ORR ground water, pointing to the ecological importance of phages in these microbial communities.
To our knowledge, this is the first study to identify bacteriophage distribution in a groundwater ecosystem. These findings shed light on bacteriophage prevalence and distribution across metal-contaminated and background sites, and this is the first study to actually use bacterial WGS (whole genomse sequences) from strains isolated from the same environment along with publicly known NCBI WGS. Certain phage-encoded genes may provide an environment-specific selective advantage to their bacterial hosts, impacting the composition of the ecosystem. Our research serves as a significant step towards directed phage isolations using specific bacterial host strains to further characterize groundwater phages, their lifecycles, and their effects on groundwater microbiome and biogeochemistry.
This study leveraged a previously gathered plasmidome sequence data from ground water samples of this unique environment. By implementing additional tools such as VirSorter developed by the DOE Joint Genome Institute, several viral genomes could be identified. Further, by using available whole genome sequence (WGS) for isolates from this environment, potential hosts bacteria could be proposed for these viruses. Both the plasmidome and WGS data are available via the DOE Knowledgebase (KBase narrative: https://kbase.us/n/63776/35).
