Simulating subsurface environments reveals complexities of microbial interactions.
The Science
Researchers in the Institute for Systems Biology’s Baliga Lab examined model organisms from two classes of microbes whose interaction converts over a gigaton of carbon to methane each year. They published their work in Nature Communications. Senior research scientist Jacob Valenzuela, who led the project, found that gene mutations selected for over a relatively short timeframe in the two microbes –Desulfovibrio vulgaris (Dv) and Methanococcus maripaludis (Mm)– led to distinct functions, specifically ecotypes, or sub-populations, that are more specialized to either an attached or floating lifestyle.
The Impact
This experiment shows how few mutations are needed to generate ecotypes of microorganisms specialized to new environmental niches. Findings show that the interplay between ecotypes of diverse microbes across space and time can dramatically improve the productivity of the entire microbial community. This study also provides insight into the ability of microbial communities to adapt, specialize, and collaborate to influence the biogeochemical cycling of gigatons of major elements like carbon, nitrogen, and phosphorus that are essential for sustaining life on Earth.
Summary
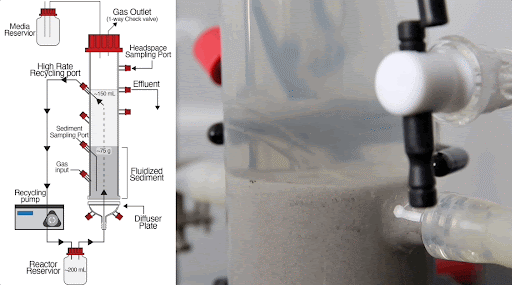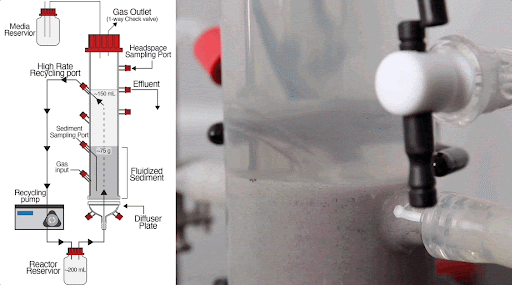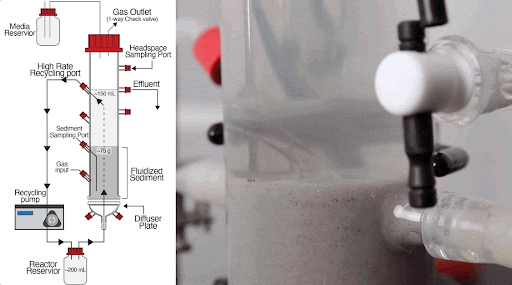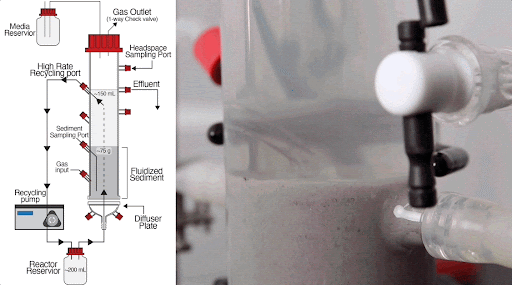
The team built a customized fluidized bed reactor to simulate the soil subsurface, where methane-producing microbial communities are typically found. The reactor uses an up-flow current to fluidize a bed of solid particles into a suspension, similar to how soil particles are suspended in flowing groundwater in the subsurface. This type of reactor system allows researchers to examine complex microbial interactions that normally occur in the soil subsurface in a controlled laboratory experiment.
Using this system, they discovered that just a handful of mutations arising within 300-1,000 generations (i.e., months in the lab) of growing Dv and Mm together generated new ecotypes of each organism, with each ecotype specialized to carry out specific roles in the simulated subsurface environment.
New ecotypes of Dv that had specialized to survive as biofilm attached to a sediment particle utilized more lactate, generating a byproduct of molecular hydrogen (H2) that nearby ecotypes of attached Mm could only partially utilize. The unused H2 was then scavenged by free-flowing Mm ecotypes and used for producing a large amount of methane.
Contact

Jacob J. Valenzuela, Sr. Research Scientist
Institute for Systems Biology
Jake.Valenzuela@isbscience.org
Funding
This study is funded by the Genome Sciences Program of the Office of Biological and Environmental Research to ENIGMA – Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Science Focus Area Program at Lawrence Berkeley National Laboratory that is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research.
Publication
Valenzuela, J.J.; S.R. Christinal Immanuel, J. Wilson, S. Turkarslan, M. Ruiz, S.M. Gibbons, K.A. Hunt, M. Auer, M. Zemla, D.A. Stahl and N.S. Baliga (2024) Origin of biogeographically distinct ecotypes during laboratory evolution. Nature Communications. [DOI]:10.1038/s41467-024-51759-y OSTI:2438359
