ENIGMA researchers at Lawrence Berkeley Lab and the University of Missouri systematically used high-throughput genetics to fill gaps in amino acid biosynthesis pathways. This explains how bacteria can grow on their own, in contrast to widespread speculation that bacteria cross-feed amino acids among community members.
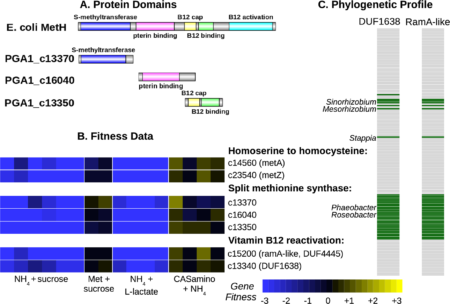
 According to Morgan Price, for many bacteria with sequenced genomes, we do not understand how they synthesize some amino acids. This makes it challenging to reconstruct their metabolism, and has led to speculation that bacteria might be cross-feeding amino acids. We studied heterotrophic bacteria from 10 different genera that grow without added amino acids even though an automated tool predicts that the bacteria have gaps in their amino acid synthesis pathways. Across these bacteria, there were 11 gaps in their amino acid biosynthesis pathways that we could not fill using current knowledge. Using genome-wide mutant fitness data, we identified novel enzymes that fill 9 of the 11 gaps and hence explain the biosynthesis of methionine, threonine, serine, or histidine by bacteria from six genera. We also found that the sulfate-reducing bacterium Desulfovibrio Vulgaris synthesizes homocysteine (which is a precursor to methionine) by using DUF39, NIL/ferredoxin, and COG2122 proteins, and that homoserine is not an intermediate in this pathway. Our results suggest that most free-living bacteria can likely make all 20 amino acids and illustrate how high-throughput genetics can uncover previously-unknown amino acid biosynthesis genes.
According to Morgan Price, for many bacteria with sequenced genomes, we do not understand how they synthesize some amino acids. This makes it challenging to reconstruct their metabolism, and has led to speculation that bacteria might be cross-feeding amino acids. We studied heterotrophic bacteria from 10 different genera that grow without added amino acids even though an automated tool predicts that the bacteria have gaps in their amino acid synthesis pathways. Across these bacteria, there were 11 gaps in their amino acid biosynthesis pathways that we could not fill using current knowledge. Using genome-wide mutant fitness data, we identified novel enzymes that fill 9 of the 11 gaps and hence explain the biosynthesis of methionine, threonine, serine, or histidine by bacteria from six genera. We also found that the sulfate-reducing bacterium Desulfovibrio Vulgaris synthesizes homocysteine (which is a precursor to methionine) by using DUF39, NIL/ferredoxin, and COG2122 proteins, and that homoserine is not an intermediate in this pathway. Our results suggest that most free-living bacteria can likely make all 20 amino acids and illustrate how high-throughput genetics can uncover previously-unknown amino acid biosynthesis genes.
Price, M.N.; G. M. Zane, J. V. Kuehl, R. A. Melnyk, J. D. Wall, A. M. Deutschbauer, A. P. Arkin (2018) Filling Gaps in Bacterial Amino Acid Biosynthesis Pathways with High‐throughput Genetics. PLOS Genetics.
DOI:10.1371/journal.pgen.1007147
Results can be viewed http://fit.genomics.lbl.gov