Adam Arkin, lead scientist, along with ENIGMA team members Bradley Biggs , a postdoctoral researcher in the Arkin lab, and Mingfei Chen, a postdoctoral researcher in the Chakraborty lab, recently participated in a virtual Research Slam promoting science related to the BioEPIC building. https://bioepic.lbl.gov/slam/
Results: Mingfei Chen (ENIGMA) took first place, Mo Ombadi (Watershed Function) second, and Bradley Biggs (ENIGMA) tied for honorable mention with Avery Roberts (m-CAFEs).
 Mingfei Chen, Postdoctoral Researcher- reigning BioEPIC Slam Champion
Mingfei Chen, Postdoctoral Researcher- reigning BioEPIC Slam Champion
“Journey to the Depths: A Subterranean Society of Microbial Biofilms“
Mingfei is a postdoc working with Romy Chakraborty and has been at Berkeley Lab for almost a year. His research focuses on the survival of the environmental microbes. He comes from a geology background, and is interested in how to use both geological and microbial records to better understand the Earth’s history. He started playing piano at the age of 6, and still enjoys it now.
Mingfei’s Flashtalk: Imagine a busy city deep below the earth’s surface, teeming with life and activity. But instead of skyscrapers and cars, this city is made up of tiny microorganisms, also known as subsurface microbes. However, unlike the surface environments, such as the soil, lake and ocean, subsurface environments are usually more harsh, with no sunlight and low nutrients. So it’s challenging for microbes to live there, but studies have been showing that subsurface microbes might actually contain up to 70% of the planet’s total microbial biomass. So how can those tiny bugs survive and thrive in those challenging environments? One strategy is to form biofilms. Think of biofilms like a city’s fortress. Just like how a fortress provides shelter, protection, and resources for the people living inside, biofilms provide a safe haven for microorganisms to thrive in harsh environments. The slimy matrix that biofilms create can act like a protective wall, shielding the microorganisms from toxic chemicals, high pressures, and low nutrients. Additionally, the microorganisms within the biofilm can work together to share resources with each other, just like how a city’s inhabitants cooperate and communicate within the fortress. Although biofilms are very commonly studied in the medical and industrial world such as oral biofilm and oil pipes, information on environmental biofilms and specifically, in the deep subsurface is absent. We believe that if microbes are organized in a biofilm, they can tolerate the extreme conditions better, and even help to trap the migrations of some of the toxic metals such as uranium. In my research, we are looking at a certain group of microbes, genus Rhodanobacter, which are abundant in our highly contaminated field sites and are resilient to environmental stresses. When looking at if they can form biofilms that protect them from extreme environmental stresses, such as toxic metals and low pH, we uncovered that certain metals, such as aluminum and manganese, actually stimulate the formation of these slimy biofilms. We’re now working with a team of BioEpic researchers to uncover the secret genes that play key roles in the biofilm forming process, and using advanced microscopy to take an up-close look at how these slimy communities are built. Our next step is to put these bugs back into the field, and observe how they behave in their natural habitat. Overall, in environments where life is tough and survival is key, biofilms show us how important it is to work together and adapt to the challenges we face.
Bradley Biggs, Postdoctoral Researcher –honorable mention
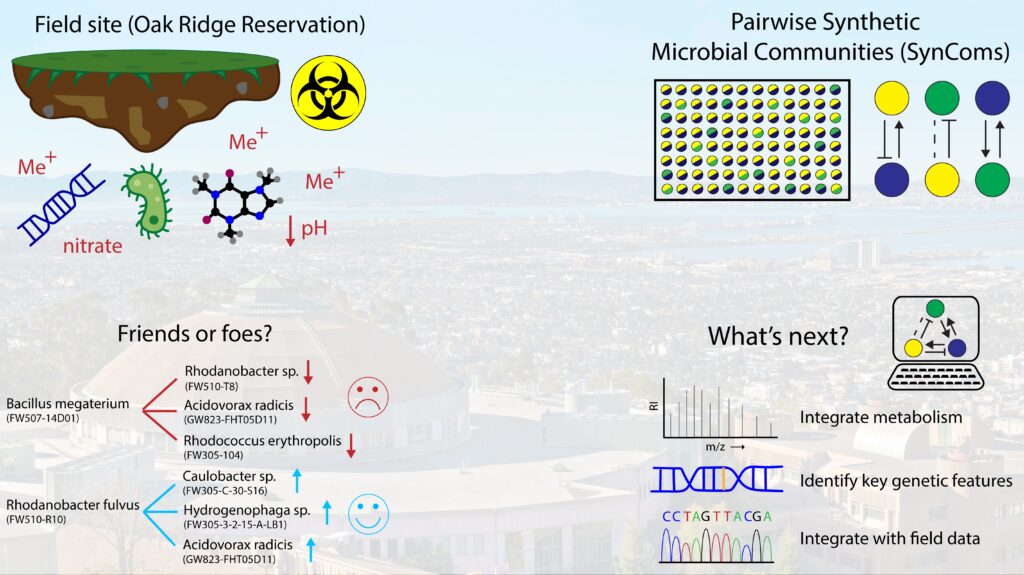
“Friends or foes forged under fire? Using synthetic microbial communities to understand field interactions”
Bradley is a postdoc working with Adam Arkin and has been at Berkeley Lab for a year and a half. His research focuses on synthetic microbial communities. He comes from a chemical engineering background, and is interested in how we could eventually apply what we learn in ecology to engineering. Bradley enjoys the outdoors, but doesn’t recommend overnight camping at Point Reyes in January if your sleeping bag isn’t rated for it.
Bradley’s Flashtalk: Our big picture goal with ENIGMA is “predictive microbial ecology”.
What that means is we want to take things that are easy to measure from the environment (like DNA) and from that to predict the function of a microbial community.We want to know what bacteria, fungi, and archaea are doing and how they are interacting with each other. Eventually we want to be to the point where that measurement could occur in many different environments from the carpet in your office or home, in the ocean, or at a wastewater facility.
Right now, for ENIGMA, we are looking at a soil and subsurface environment in Oak Ridge, Tennessee. This site is interesting because it contains historic contamination from the improper disposal of hazardous waste. This led to high nitrate concentration, heavy metal contamination, and the area to be quite acidic. Interestingly, some microbes can not only survive this harsh environment, but have the potential to remediate as well. A lot of work remains to be done to close the gap between DNA measurement and predicting a microbiome’s function; and folks across ENIGMA are working on addressing different elements of this.
My project is trying to understand how the microbes interact with one another. To test this, I create synthetic microbial communities or SynComs from microbes taken from the field and grow them together in the lab to see how they interact. These tests are done pairwise in simple 96-well plates, and after the strains are grown together we measure to find out how much of each strain is present to determine if the interaction was positive, negative, or neutral. Tests are run in multiple different conditions, including those that mimic some of the hazards from the site like heavy metals and acidic conditions.
From our tests we’ve begun to identify strains that are both good and bad neighbors (or cooperative or competitive partners). One example is Bacillus megaterium acting as a competitor with several strains and Rhodanobacter fulvus as a good partner. Something that stands out is the same Acidovorax strain that is negatively impacted by the Bacillus strain is helped by the Rhodanobacter strain. In addition, all three strains shown to be helped by the Rhodanobacter species were helped in the metals and acid condition. On their own, these strains can’t grow in that environment, but with the Rhodanobacter they do.
Meaning it isn’t just a friendly neighbor but actually can help rescue them in a tough environment!
As we continue to study these strain interactions, we want to use computational techniques to incorporate the metabolisms of each of these strains (meaning what things they eat and produce) into our understanding of their interactions. Additionally, we want to look for features on their genome that could be predictors of survival in harsh environments and potentially of new successful partnerships.
Lastly, we want to take lab discovered interactions and integrate it with the measurements from the field site to see if it helps us understand what we’ve observed there. We hope that this information can not only help us begin to make better predictions of microbiome function, but help us engineer microbial communities for other applications.