For many microbes, we know little about them beyond their genome sequences. In principle, we could use genome sequences to predict microbes’ traits, such as which carbon sources they can eat, but first we need to identify more of the genes involved. We built an automated tool, GapMind, to annotate the transporters and enzymes for utilizing 62 common carbon sources. Then we used GapMind to identify gaps: transporters or enzymes that should be present, to explain how a bacterium uses a carbon source, but could not be found in the genome.
By comparing these gaps to large-scale genetic data for 29 bacteria, we identified hundreds of novel transporters and enzymes, and a new metabolic pathway for consuming glucosamine. When we added these novel genes to GapMind, its results for diverse bacteria and archaea improved significantly. However, there are still too many gaps in our knowledge to predict these traits. To do that, we’ll need large-scale data from diverse microbes about what carbon sources they can use, and also more genetic data.
We also discovered some truly novel enzymes that perform reactions that were not previously known (like NagX in the figure), or which were known, but had not been linked to a gene before. These discoveries might help engineers modify bacteria to make useful compounds.
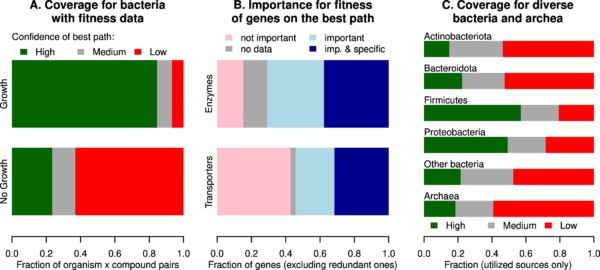
To discover novel catabolic enzymes and transporters, we combined high-throughput genetic data from 29 bacteria with an automated tool to find gaps in their catabolic pathways. GapMind for carbon sources automatically annotates the uptake and catabolism of 62 compounds in bacterial and archaeal genomes. For the compounds that are utilized by the 29 bacteria, we systematically examined the gaps in GapMind’s predicted pathways, and we used the mutant fitness data to find additional genes that were involved in their utilization. We identified novel pathways or enzymes for the utilization of glucosamine, citrulline, myo-inositol, lactose, and phenylacetate, and we annotated 299 diverged enzymes and transporters. We also curated 125 proteins from published reports. For the 29 bacteria with genetic data, GapMind finds high-confidence paths for 85% of utilized carbon sources. In diverse bacteria and archaea, 38% of utilized carbon sources have high-confidence paths, which was improved from 27% by incorporating the fitness-based annotations and our curation. GapMind for carbon sources is available as a web server (http://papers.genomics.lbl.gov/carbon) and takes just 30 seconds for the typical genome.
Price M.N.; AM. Deutschbauer and A.P. Arkin (2022) Filling Gaps in Bacterial Catabolic Pathways with Computation and High-throughput Genetics. PLoS Genetics. [DOI]:10.1371/journal.pgen.1010156 {PMID}:35417463 OSTI:1862968
Related Links
GapMind: Automated annotation of catabolism of small carbon sources
Contact
Morgan Price
Lawrence Berkeley Lab
mnprice@lbl.gov