iDIRECT could help scientists better understand biological systems by disentangling direct from indirect relationships in association networks.
The Science
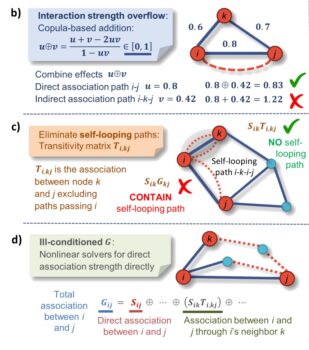
In complex dynamic systems, various components interact with one another through exchange of materials, energy, and information. Such systems can be represented as networks. When reconstructing networks from large scale datasets, particularly for biological systems, one major challenge is that networks reconstructed with statistical approaches contain both direct and indirect associations, resulting in biased network structures with spurious links and inaccurate weights. ENIGMA researchers developed a general approach to disentangle direct from indirect relationships in association networks. The tool is called iDIRECT (Inference of Direct and Indirect Relationships with Effective Copula-based Transitivity).
The Impact
By tackling several mathematical challenges, iDIRECT provides a conceptual framework to disentangle direct and indirect relationships in association networks. The framework will greatly enhance our capability to discern network interactions in various complex systems. It will also allow scientists to address research questions which could not be approached previously.
Summary
ENIGMA researchers present iDIRECT as a novel framework for quantitatively inferring direct dependencies in association networks. Using copula-based transitivity, iDIRECT ameliorates several challenging mathematical problems. With simulation data as benchmark examples, iDIRECT showed high prediction accuracies. Application of iDIRECT to reconstruct gene regulatory networks in Escherichia coli also revealed considerably higher prediction power than the best performing approaches in the DREAM5 Network Inference Challenge. In addition, applying iDIRECT to highly diverse grassland soil microbial communities in response to climate warming showed that the iDIRECT-processed networks were significantly different from the original networks, with considerably fewer nodes, links, and connectivity, but higher relative modularity. Further analysis revealed that the iDIRECT-processed network was more complex and more robust. As a general approach, iDIRECT has great advantages for network inference, and it should be widely applicable to infer direct relationships in association networks across diverse disciplines in science and engineering.
Contact

Jizhong Zhou, George Lynn Cross Research Professor; Director, Institute for Environmental Genomics (IEG)
Department of Microbiology & Plant Biology, University of Oklahoma
jzhou@ou.edu

Naijia Xiao, Research Scientist
Institute for Environmental Genomics (IEG)
Department of Microbiology & Plant Biology, University of Oklahoma
naijia.xiao@ou.edu
Funding
This study was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (DOE-BER) Genomic Science program; ENIGMA – Ecosystems and Networks Integrated with Genes and Molecular Assemblies, a Science Focus Area Program at Lawrence Berkeley National Laboratory, supported by DOE-BER; and the U.S. National Science Foundation (NSF).
Publications
Xiao N., Zhou A., Kempher M.L., Zhou B.Y., Shi Z.J., Yuan M., Guo X., Wu L., Ning D., Van Nostrand J., Firestone M.K., Zhou J. “Disentangling direct from indirect relationships in association networks.” Proceedings of the National Academy of Sciences 119, no. 2 (2022). [DOI: 10.1073/pnas.2109995119]
Related Links
Science Magazine: iDIRECT network framework developed at the University of Oklahoma could help scientists better understand biological systems
