
Gushgari-Doyle, S; L.M. Lui, T.N. Nielsen, X. Wu, R.G. Malana, F.L. Poole II, M.W.W. Adams, A.P. Arkin, R. Chakraborty (2022) Genotype to ecotype in niche environments: Adaptation of Arthrobacter to carbon availability and environmental conditions. ISME Comm. [DOI]:10.1038/s43705-022-00113-8 OSTI:1860268
Different niche micro-environments can affect the structure and function of microbial communities. Terrestrial subsurface environments exhibit specific biogeochemical conditions associated with depth, often resulting in such distinct environmental niches, as is also observed at the ENIGMA field site at ORR. A team of ENIGMA scientists examined seven strains of Arthrobacter across a sediment core, finding that the strains varied functionally and genomically depending on the ecotype from which they were isolated.
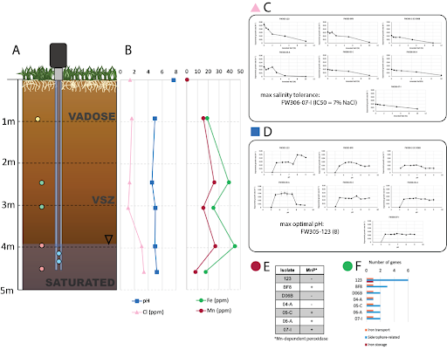
The fact that environmental niches affect genotype, phenotype, structure, and function has been researched more extensively between environments with more extreme differences, such as between freshwater and hot springs. This study focuses on microniches that are physically adjacent but diverse; subsurface environments can change significantly across a minimal amount of space due to geochemistry, carbon availability, and nearby energy sources. It is also less well-known how organisms of the same genus may adapt to microniches.
In this study, Arthrobacter strains were found to be capable of adapting to the microniches within the sediment core. For instance, increased salinity tolerance was identified in a strain present where salt concentrations were the highest. Arthrobacter also has a volatile accessory genome and an open pangenome, suggesting it may be a good model organism for studying rapid adaptation to environmental niches.
This work has also been profiled by the Earth and Environmental Sciences Area and Berkeley Lab Biosciences.

Contact
Romy Chakraborty <rchakraborty@lbl.gov>
Environmental Atlas Science Co-Lead
