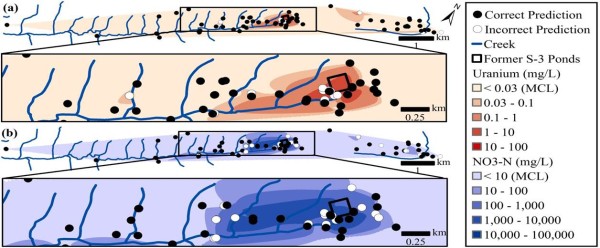
ENIGMA scientists show that DNA from natural bacterial communities can be used as a quantitative biosensor to accurately distinguish unpolluted sites from those contaminated with uranium, nitrate, or oil.
- Bacterial communities yield a predictable response to environmental constraints.
- Bacterial systems can be used forensically to report the occurrence of environmental perturbations.
- Bacterial systems have the potential to delineate useful stewardship strategies
Statistical analysis of DNA from natural microbial communities can be used to accurately identify environmental contaminants, including uranium and nitrate at a nuclear waste site. In addition to contamination, sequence data from the 16S rRNA gene alone can quantitatively predict a rich catalog of 26 geochemical features collected from 93 wells with highly variable geochemistry. As a whole, these results indicate that ubiquitous, natural bacterial communities can be used as in-situ environmental sensors that respond to, and capture, perturbations caused by human impacts and natural extremes. These in-situ biosensors rely on environmental selection rather than directed engineering. So this approach could be rapidly deployed and scaled as sequencing technology continues to become faster, simpler, and less expensive.
More immediately, by demonstrating the rich geochemical information captured by bacterial communities, this work supports the view that bacterial communities yield a predictable response to environmental constraints. This association between community and constraint means that bacterial systems can be used as sentinels to report the impacts of environmental perturbations, and potentially to delineate useful stewardship strategies.
More stories related to this paper can be found: