
- Direct PCR method exhibits an overall efficiency = conventional DNeasy PowerSoil protocol
- 1,600X CHEAPER ($0.34 / 96 samples) + 10X SIMPLER/FASTER (15 min hands-on time /96 samples)
- Fully automated + compatible with small-volume samples
Song, F.; J.V. Kuehl, A. Chandran, A.P. Arkin (2021) A cheap, simple and automation-friendly direct PCR approach for bacterial community analysis. mSystems [doi]:10.1128/mSystems.00224-21 OSTI:1822977
Bacterial communities in water and soil play an essential role in environmental ecology and human health. It is critical to know the microbial composition, dynamics, and interactions. Sequencing-based pipeline is the most popular way to analyze microbial communities. However, the genomic DNA extraction is time-consuming, expensive, and often biased based on the types of species present. This study reports a direct PCR approach to replace the tedious gDNA extraction. This direct PCR approach exhibits a comparable efficiency with the traditional methods, but 1,600 times less expensive ($0.34 for 96 samples) and 10 times simpler (15 min hands-on time for 96 samples).
Understanding bacterial interactions and assembly in complex microbial communities using 16S rRNA sequencing normally requires a large experimental load. The current pipeline is costly, time-consuming, labor-intensive, and not amenable to miniaturization by droplets or 1,536-well plates. This direct PCR method could potentially solve these problems. This simple, cost-effective, and automation-friendly direct-PCR-based 16S rRNA sequencing method allows us to study the community dynamics, microbial interaction, and assembly of various complex microbial communities in a high-throughput fashion.
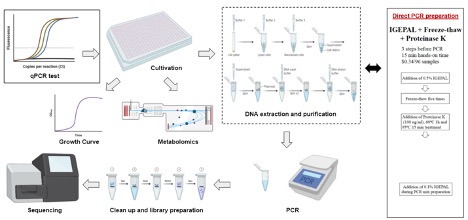
Bacterial communities play an essential role in environmental ecology and human health. PCR-based amplicon analysis is a fundamental tool for quantifying and studying microbial composition, dynamics, and interactions. However, given the complexity of microbial communities, a substantial number of samples becomes necessary for analyses that parse the factors that determine microbial composition. A common bottleneck in performing these kinds of experiments is genomic DNA extraction, which is time-consuming, expensive, and often biased based on the types of species present. Here, we reported that a specific variant of the direct PCR method exhibits an overall efficiency comparable to that of the conventional DNeasy PowerSoil protocol in the circumstances we tested. This direct PCR method is 1,600 times less expensive ($0.34 for 96 samples) and 10 times simpler (15 min hands-on time for 96 samples) than the DNeasy PowerSoil protocol. The direct PCR method can also be fully automated and is compatible with small-volume samples, thereby permitting scaling of samples and replicates needed to support high-throughput large-scale bacterial community analysis.
Contact
Adam P. Arkin <aparkin@lbl.gov>
Professor, Department of Bioengineering, University of California, Berkeley
Senior Faculty Scientist, Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory
Technical Co-Lead, ENIGMA SFA
CEO/CSO, DOE Systems Biology Knowledgebase
