The Science
Bacillus cereus str. CPT56D-587-MTF was isolated from the highly contaminated subsurface of the Oak Ridge Reservation in Oak Ridge, TN. This site is contaminated with nitric acid and heavy metals. Using molecular analyses, we discovered that this strain is a representative of a highly abundant species at the site. By conducting laboratory studies and analyzing the genome of this isolate, we discovered adaptations that have allowed this strain to survive and flourish in its highly stressful environment. We found that mobile genetic elements (pieces of DNA capable of autonomous replication and moving around and between genomes) have been important in the evolution of this strain.
The Impact
Industrial waste sites are frequently co-contaminated with heavy metals and nitrate. While nitrate can be removed from the site by native microorganisms through a process known as nitrate-respiration, heavy metal co-contaminants can inhibit this process. It is important to determine how heavy metals impact the physiology and evolution of key nitrate-respiring microorganisms at the site. This study will aid researchers in developing predictive models for understanding microbial processes at contaminated sites.
Summary
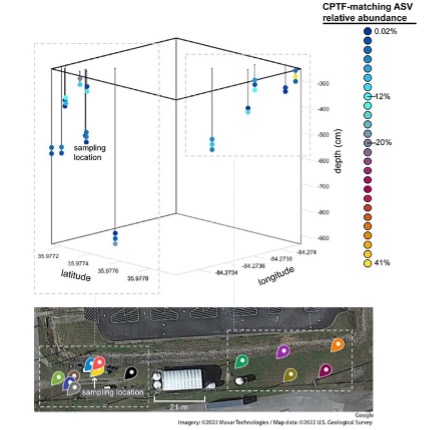
This research used a combination of pangenomic, community analysis, and laboratory phenotyping approaches to understand how dominant microorganisms inhabit their unique ecological niche within a highly contaminated subsurface environment. The US Department of Energy (DOE) Oak Ridge Reservation (ORR) in Tennessee contains a subsurface site that is contaminated by multiple heavy metals and nitric acid. This contamination is due to the legacy of waste discharge from uranium processing operations that occurred at the on-site Y-12 National Security Complex into waste disposal ponds between 1951 and 1983. This waste leached from the disposal ponds into the surrounding subsurface. The near-source groundwater and soil is highly acidic and contaminated with high levels of nitrate, uranium and other metals. We conducted a community survey of the sediments in a site immediately adjacent to the former waste disposal ponds. We found that the most dominant species within these sediments was Bacillus cereus. To investigate the adaptations of this species to the various stressors present in its environment, we isolated a representative strain from this site, B. cereus str. CPT56D-587-MTF (strain CPTF).
We obtained a complete genome sequence of strain CPTF that was significantly longer and has greater plasmid content than other reported B. cereus strains, was significantly longer and had greater plasmid content. Plasmids are important for acquisition of auxiliary genes important for survival in fluctuating environments. Our analyses revealed multiple genes for heavy metal resistance and acid tolerance across the eight CPTF plasmids. This strain also had evidence of expansion of transposable elements. Transposable element duplication and mobilization allow for rapid genomic change during periods of stress. Further pangenome analysis revealed that certain features important for survival in the ORR subsurface such as nitrate respiration enzymes and heavy metal efflux pumps are near-ubiquitous among B. cereus species. This suggests that these traits may have facilitated the proliferation of strain CPTF at the site.

Contact
Michael Adams
University of Georgia
adamsm@uga.edu
Publications
Goff JL, Szink EG, Thorgersen MP, Putt AD, Fan Y, Lui LM, Nielsen TN, Hunt KA, Michael JP, Wang Y, Ning D, Fu Y, Van Nostrand JD, Poole II FL, Chandonia, J-M, Hazen TC, Stahl DA, Zhou J, Arkin AP, Adams MWW. 2022. Ecophysiological and genomic analyses of a representative isolate of highly abundant Bacillus cereus strains in contaminated subsurface sediments. Environmental Microbiology. https://doi.org/10.1111/1462-2920.16173
Goff JL, Lui LM, Nielsen TN, Thorgersen MP, Szink EG, Chandonia J-M, Poole II FL, Zhou J, Hazen TC, Arkin AP, Adams MWW. 2022. Complete genome sequence of Bacillus cereus strain CPT56D-587-MTF, isolated from a nitrate- and metals-contaminated subsurface environment. Microbiology Resource Announcements. https://doi.org/10.1128/mra.00145-22
