Lui, Lauren M.; T.N. Nielsen, A.P. Arkin (2021) A method for achieving complete microbial genomes and better-quality bins from metagenomics data. PloS Computational Biology [DOI]:1371/journal.pcbi.1008972 OSTI:1788019
A Method for Achieving Complete Microbial Genomes and Improving Bins From Metagenomics Data
Assembling complete microbial genomes from short read metagenomics data is difficult, but the method “Jorg” helps semi-automate the process.
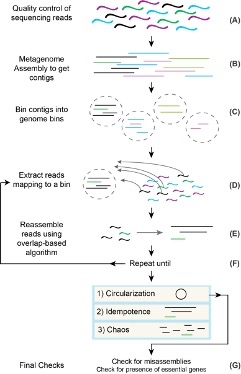
Since we cannot culture many microorganisms that are found in the environment, animals, and the human body, scientists rely on shotgun metagenomics sequencing to study their genomes and infer what they can do. However, shotgun metagenomics often only provides partial genomes due to limitations of available sequencing technology and computational tools for genome assembly. We present a semi-automated method called Jorg that can be used to improve and eventually complete microbial and viral genomes from metagenomics data.
At the time of this study, there were only ~100 known complete bacterial genomes from metagenomes from ~30 other studies. Bacterial genomes are considered complete if they are circular and do not have misassemblies. We circularized 36 bacterial genomes and two megaphage (viral) genomes, which is much more than what any single study had previously done and represents a 36% increase in available circularized bacterial genomes. This method provides a way for scientists to finish genomes from samples where we cannot culture the microbes and provide high-quality genomes for comparative genomics studies.
In our manuscript we describe a semi-automated method called Jorg that facilitates recovery of complete (circular) archaeal, bacterial, and viral genomes from metagenomics data and that also provides checks for misassemblies. As a proof-of-concept we circularized 36 bacterial genomes and two megaphage genomes. For comparison, there are only ~100 known circularized bacterial genomes from metagenomes from ~30 other studies. We also demonstrate findings that illustrate the utility of circularizing genomes by discovering new biological patterns in Candidate Phyla Radiation species. High-quality circularized genomes produced using this tool also can be used as scaffolds to improve future genome assemblies and as data to improve identification of species in microbiomes.
Related Links
Contact
Adam P. Arkin
Professor, Department of Bioengineering, University of California, Berkeley
Senior Faculty Scientist, Environmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory
CEO/CSO, DOE Systems Biology Knowledgebase
aparkin@lbl.gov
