Nitrate tolerance mechanisms in D. vulgaris revealed by experimental evolution.
Wu, B., Liu, F., Zhou, A. et al., “Experimental evolution reveals nitrate tolerance mechanisms in Desulfovibrio vulgaris.” ISME J 14, 2862-2876 (2020). [doi: 10.1038/s41396-020-00753-5]
The Science
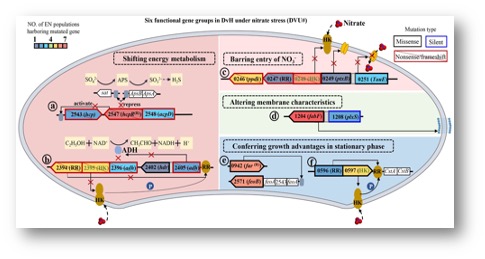
To uncover the nitrate tolerance mechanisms, twelve Desulfovibrio vulgaris populations were propagated under nitrate stress for 1000 generations. The evolved populations showed increase nitrate tolerance. DNA sequencing revealed frequently mutated six groups of genes, including nitrosative stress response genes, nitrogen regulatory protein C family genes, nitrate cluster genes, fatty acid synthesis genes, iron regulatory genes, and two-component system genes LytR/LytS. Functions of these genes in nitrate tolerance were confirmed by mutant analysis. The loss-of-function mutant of the individual gene showed increased nitrate tolerance, while the introduction of the gene into the mutant restored the wild-type like nitrate tolerance phenotype.
The Impact
Understanding the microbial adaption mechanisms to environmental stresses is a central topic in biology. Sulfate-reducing bacteria (SRB) use sulfate as the terminal electron acceptor and play important roles in the biogeochemical cycling of sulfur, carbon, nitrogen, and metals. Nitrate, an energetically more favorable electron acceptor, usually co-exist with sulfate in the natural environment. Although low nitrate does not inhibit sulfate-reduction, it is unknown how SRB that lack the nitrate reductase, like DvH, survives in an environment with elevated nitrate. Here, experiment evolution of DvH under elevated nitrate was conducted, analysis of the evolved populations and mutants revealed the possible nitrate adaptation mechanisms.
Summary
Elevated nitrate in the environment inhibits sulfate reduction in sulfate-reducing bacteria (SRB). Several SRB may respire nitrate to survive, but how SRB that lack nitrate reductase survives under elevated nitrate conditions remains elusive. To uncover nitrate adaptation mechanisms, 12 populations of Desulfovibrio vulgaris Hildenborough (DvH), a model SRB, were propagated under elevated NaNO3 for 1000 generations. Nitrate-evolved (EN) populations showed significantly (p < 0.05) increased nitrate tolerance, and whole-genome resequencing identified 119 new mutations in 44 genes, among which six gene groups showing high mutation frequencies at the population level. For instance, a high frequency of nonsense or frameshift mutations in nitrosative stress response genes, nitrogen regulatory protein C family genes, and nitrate cluster genes. Mutants with loss-of-functions of NRC and NSR showed increased nitrate tolerance. In addition, a high frequency of missense mutations was found in genes involved in the fatty acid synthesis, iron regulation, and two-component system genes lytR/lytS known to be responsive to multiple stresses. These mutations could increase nitrate tolerance by regulating energy metabolism, barring entry of nitrate into cells, altering cell membrane characteristics. Overall, this study advances our understanding of nitrate tolerance mechanisms and has important implications for linking genotypes to phenotypes in DvH.
Contact
Jizhong Zhou
Professor, Department of Microbiology and Plant Biology, University of Oklahoma
jzhou@ou.edu, phone: 405-325-6073
Zhili He
Professor, Sun Yat-sen University
