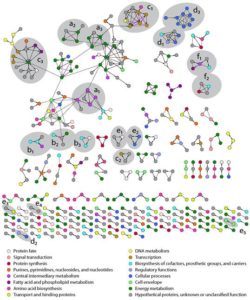
The first demonstration of a highly accurate, genome-wide method for detecting stable protein-protein interactions, without the requirement for genetic manipulation of the organism (prior tagless methodologies have false discovery rates of >85%).
Combined with our study of protein complexes published in February 2016, which was based on affinity purification coupled with mass spectroscopy (AP-MS), this experiment yielded a more complete picture of the stable protein interactome of the model sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough, further enabling evidence-based annotation of gene function.
