 ENIGMA researchers at University of Tennessee Knoxville examined differences in diversity between extraction techniques.
ENIGMA researchers at University of Tennessee Knoxville examined differences in diversity between extraction techniques.
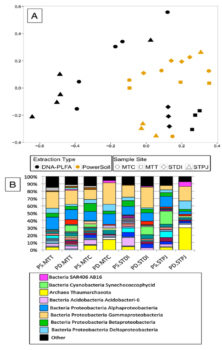
- Lipid/DNA co-extraction from one sample is attractive in limiting biases associated with microbial community analysis from separate extractions. We sought to enhance established co-extraction methods and use high-throughput 16S rRNA sequencing to identify preferentially extracted taxa from co-extracted DNA.
- Co-extraction results in low DNA yields and distinct community structure changes.
- Anecdotally, many people (including our researchers) have tried this technique and failed over the past decade though nothing has ever been published about using co-extraction. This study documents the method, in order to set the context for future work in the area.
