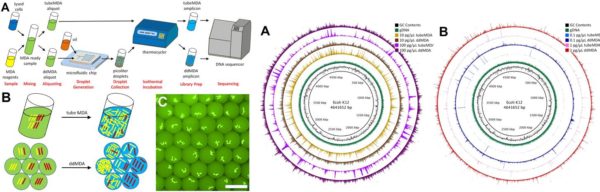
- The ddMDA technique enabled significantly lower bias and non-specific amplification than conventional MDA thus achieving more uniform coverage of amplification over the entire genome
- This technique can be a powerful tool for genomic studies where DNA samples are limited such as single cells, microaggregates, and uncultured microbes from many different environments
Digital Droplet Multiple Displacement Amplification (ddMDA) for whole-genome sequencing of limited DNA samples (2016) Rhee, M., Light, Y.K., Meagher, R.J., and Singh, A.K. Plos One [doi]10.1371/journal.pone.0153699
